18 Apr 2024
If you are having problems pushing commits from Jetstream after you have modified you repo on Github.com or an another computer, you may need to update your git preferences on your Jetstream instance.
Symptom
You try to push and get an error saying that you need to pull:
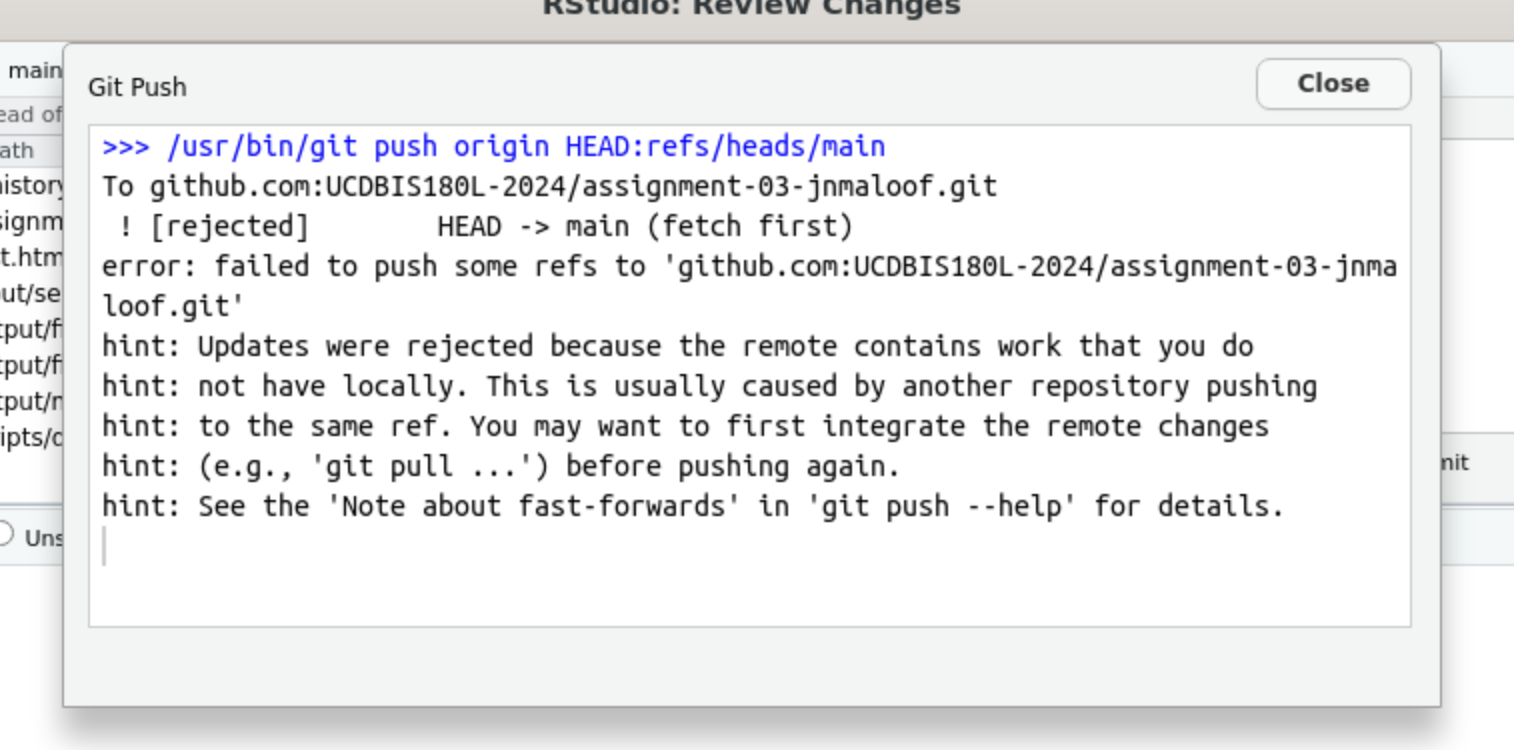
Then you try to pull, but you get another error:
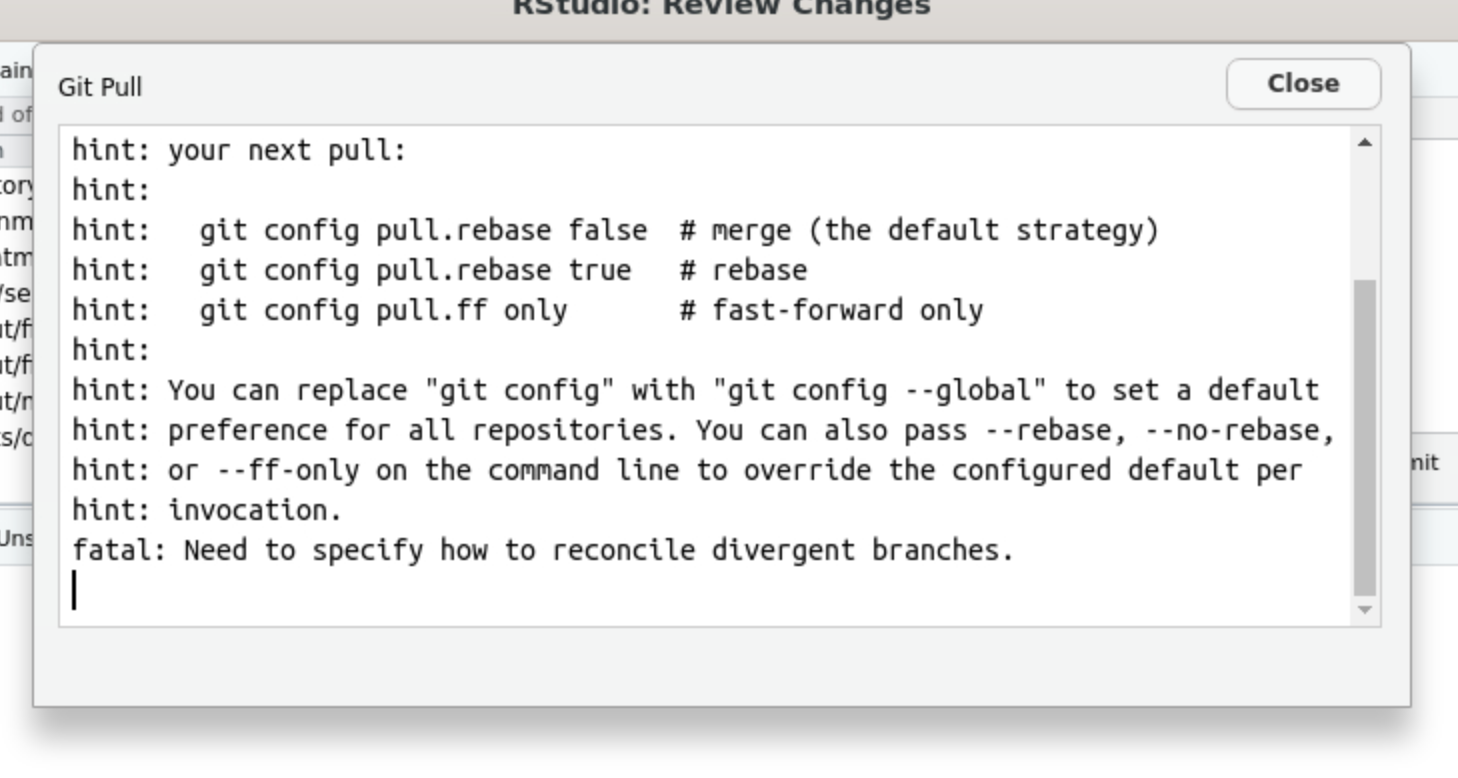
Fix
Open the Linux terminal and type:
git config --global pull.rebase false
Then go back to RStudio, pull, and then push.
23 Feb 2024
Dear Students,
Welcome to BIS180L. I am looking forward to teaching you in this class. This class will give you hands-on experience with bioinformatics data analysis.
Class will meet in person in TLC 2216
Pre-recorded Lectures
For the most part I will provided lectures in a pre-recorded format. This will allow the best use of our time together in the classroom. Pre-recorded lectures will be available at the latest at 9AM the day before the lecture/lab time. You will need to watch them in advance and answer embedded quiz questions via playposit. Due date for the quiz is 9AM the day of lecture. This give me and the TA time to review questions before class.
There is no video due for the April 02 class
Class meeting time
Even though lectures are pre-recorded, we will still meet at 1:10 PM.
In class
We will devote the beginning of each class / lab to questions on the lecture material. You will then work in small groups of two or three to work on the lab material. John (TA) and I will be available to help when you have questions.
I expect you to be in class/lab each day at 1:10. If you have a Covid or other health emergency that prevents you from coming to class, please let me know (in advance if possible).
Outside of class
You can expect to spend a fair bit of time outside of class working on the assignments. This can either be done in one of the computer labs, or on your laptop. If you are not willing to put in time outside of class this is probably not the best class for you. If you do put in the time outside of class you will be rewarded by learning a great deal about how to perform bioinformatics analyses.
23 Feb 2024
We will use Vince Buffalo’s excellent book Bioinformatics Data Skills.
This book is available online for free through the UC Davis Library (on campus or VPN connection required). UC is licensed for simultaneous access by 28 people. If that doesn’t work for you, you can buy it direct from the publisher or from amazon. As of this posting Amazon is cheaper and has used copies as well.
Additionally we will use Hadley Wickham’s also excellent book R for Data Science which is available online for free. If you would like a physical copy, here is the amazon link

